Hello! Today we'll be going through some hands-on activities to help you get familiar with how many bioinformatics tasks can be done directly from the command line.
The first thing you should do if you haven't done so is connect to the ConGen server. We'll be working exclusively in the RStudio browser interface that you should be familiar with by now, but if you have questions or problems at any point please feel free to ask! Just in case, here's an annotated picture of roughly what you should be seeing right now. If you are seeing something drastically different or something that you don't understand, let us know.
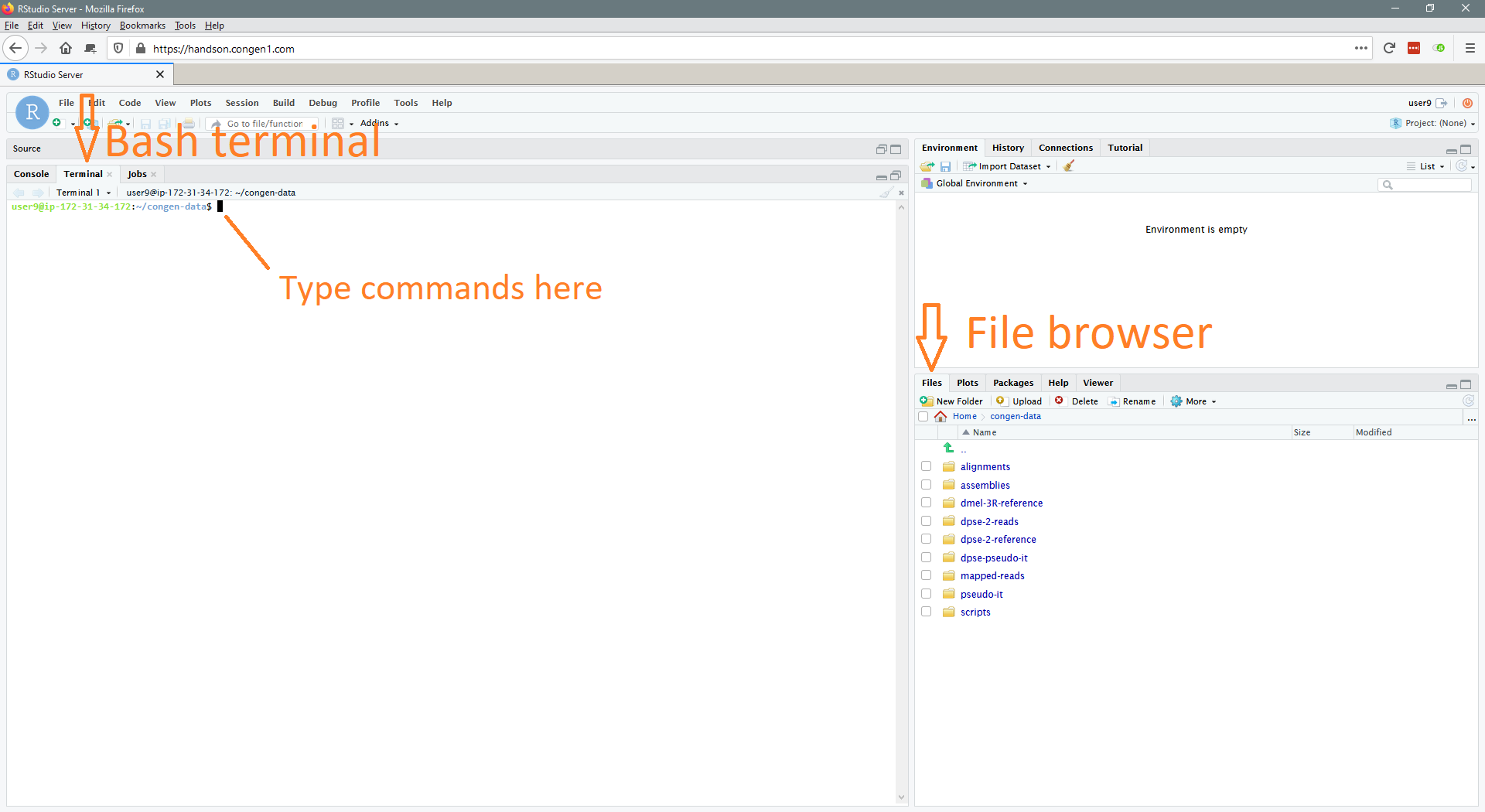
Most of our work will be done as bash commands typed in the Terminal provided by RStudio. Throughout this walkthrough, commands will be presented as follows:
this is an example commandFollowing each command will be a table that goes through and explains each part of the command explicitly:
| Command line parameter | Description |
|---|---|
| this | An example command |
| is | An example option used in the example command |
| an | An example option used in the example command |
| example | An example option used in the example command |
| command | An example option used in the example command |
The goal of providing these tables is to break-down some of the 'black box' that command line tools can sometimes feel like. Hopefully this is helpful. If not, feel free to skip over these tables when you see them!
A general convention among command-line software is to provide a help menu for programs that lists common options. These can generally
viewed from the command line with the -h option as follows:
<program> -h -or- <program> <sub-program> -hFor Linux commands, documentation is generally available with the man command (man is short for manual):
man <command>man opens a text viewer that can be navigated with the arrow keys and exited simply by typing q.
If you're ever stuck or want to know more about a program's options, try these!
Commands that you should run will have a green background. We will also provide some commands that are beneficial to see, but do not necessarily need to be run using a red background, like so:
this is an example command that won't be runAdditionally, one of the most important and often overlooked parts of bioinformatics analyses is to simply look at ones data. There will be several points where we stop to look at the output of a given program or command. When we do, a snippet of the output will be presented in the walkthrough as follows:
Here is some made up output.
Looking at your data is very important!
You can catch problems before you use the data in later analyses.Today we'll be doing some basic bioinformatics tasks from the command line. We'll get to the specifics of the data later, but for now please download the project template we've provided on github.
First, make sure you're in your home directory. If you're not, or you're not sure if you are, run the command:
cd ~| Command line parameter | Description |
|---|---|
| cd | The Linux change directory command |
| ~ | The path to the directory you want to change to. ~ is a shortcut for "the current user's home directory." |
Next, download the project repository using git:
git clone https://github.com/gwct/congen-bioinformatics.git| Command line parameter | Description |
|---|---|
| git | A cross platform program for vesrion control and syncing of software and data projects. |
| clone | The git sub-program to make download an exact copy of a repository. |
| https://github.com/gwct/congen-bioinformatics.git | The URL of the project repository. This can be found on the webpage of the repository. |
Git is very powerful software for sharing your projects and used commonly to share code and data from scientific papers,
but we won't talk about it much today other than using the clone command to download the project.
You don't need a github account to clone a repository, but you do need git installed on your computer to do so.
If you're interested in learning more about git there are a ton of guides and docs out there for you to search for. To get started, we've put together a couple of how-tos for understanding git basics here:
After the clone command completes, you should now have a folder in your home directory called congen-bioinformatics.
Make sure it's there with ls:
ls| Command line parameter | Description |
|---|---|
| ls | The Linux list directory contents command. With no other options given, this lists the contents of the current directory. |
And next change into that directory:
cd congen-bioinformatics| Command line parameter | Description |
|---|---|
| cd | The Linux change directory command |
| ~ | The path to the directory you want to change to. |
Using this project template data, we'll be performing the following tasks today:
- Talking about project organization, common commands, and text editors and work setups.
- Introducing common bioinformatics file formats.
- Using the command line to do a basic analysis of structural variation in a sample of 32 Rhesus macaques and of SNPs in 35 gray wolves.
- Time permitting, briefly touch on some next steps in developing more advanced bioinformatics skills
Now, let's move on to Project Organization
