Placing a known WGD on a phylogeny
Below are the inputs, commands, and outputs to do an analysis with GRAMPA to place a known WGD on a phylogeny. The inputs are based on simulated data. For more detailed info on the simulations check our paper.
The examples below call GRAMPA as grampa assuming it has been installed from bioconda. If you installed
from source, see usage details in the README.
Inputs
Suppose you have a set of species, of which you have evidence that a some may be the result of a polyploidization event. You also may have an idea about the parental lineages of the polyploid species. So you build a species tree of your taxa and, since species tree reconstruction programs output singly-labeled trees, you get this as the result:
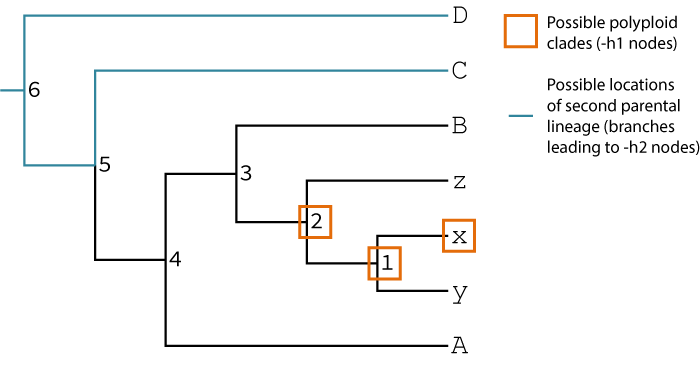
Your hypothesis is that species x, y, and z may be the result of polyploidization (but you're not sure if all of them are). The singly-labeled tree implicitly identifies one of the parental lineages of the suspected polyploids by placing the polyploid species sister to it (in this case lineage B seems to be one of the parents). You also think this tree may be the result of an allopolyploidy, and you think some lineage sister to C, D, the C,D clade, or at the root of the tree may have hybridized with some lineage related to B to form the x,y,z clade. Working with this prior knowledge GRAMPA can test your hypotheses of polyploidization.
The input files you would need are:
- Singly-labeled species tree: spec_tree_3a.tre
- Gene trees from your set of species (in this case 1000 gene trees simulated with gain and loss) : gene_trees_3a.txt
GRAMPA command
Since we have some idea of the lineages involved in the polyploidization event, we would want to limit GRAMPA's search to those lineages
with the -h1 and -h2 search parameters.
grampa -s spec_tree_3a.tre -g gene_trees_3a.txt -h1 "x 1 2" -h2 "C D 5 6" -o ex2_output -f ex2_testAbove we have specified -h1 and -h2 by using the node labels in the tree. Alternatively, we could specify
an equivalent -h1 and -h2 search by defining the labels based on the sets of tips that define them:
grampa -s spec_tree_3a.tre -g gene_trees_3a.txt -h1 "x x,y x,y,z" -h2 "C D A,x,y,z,B,C A,x,y,z,B,C,D" -o ex2_output -f ex2_testThese two commands are equivalent. The second method is slightly more cumbersome, but does not require you to have internal labels on your tree.
Although, GRAMPA can easily add internal labels to your input tree with the --labeltree command.
Outputs
The above command would create the directory ex2-output with five output files
- ex2-test-checknums.txt
- ex2-test-detailed.txt
- ex2-test-dup-counts.txt
- ex2-test.log
- ex2-test-scores.txt
Since we are trying to determine the mode of polyploidy, we are interested in the ex2-test-scores.txt file. This file contains
the total reconciliation scores for each MUL-tree considered, sorted from lowest scoring tree to highest scoring tree, and looks something like this:
mul.tree h1.node h2.node score labeled.tree
9 <2> C 5242 ((((((x+,y+)<1>,z+)<2>,B)<3>,A)<4>,(C,((x*,y*)<5>,z*)<6>)<7>)<8>,D)<9>
11 <2> <5> 7598 (((((((x+,y+)<1>,z+)<2>,B)<3>,A)<4>,C)<5>,((x*,y*)<6>,z*)<7>)<8>,D)<9>
0 NA NA 8312 ((((((x,y)<1>,z)<2>,B)<3>,A)<4>,C)<5>,D)<6>
5 <1> C 8506 ((((((x+,y+)<1>,z)<2>,B)<3>,A)<4>,(C,(x*,y*)<5>)<6>)<7>,D)<8>
10 <2> D 8618 ((((((x+,y+)<1>,z+)<2>,B)<3>,A)<4>,C)<5>,(D,((x*,y*)<6>,z*)<7>)<8>)<9>
12 <2> <6> 8845 (((((((x+,y+)<1>,z+)<2>,B)<3>,A)<4>,C)<5>,D)<6>,((x*,y*)<7>,z*)<8>)<9>
6 <1> D 8854 ((((((x+,y+)<1>,z)<2>,B)<3>,A)<4>,C)<5>,(D,(x*,y*)<6>)<7>)<8>
7 <1> <5> 8939 (((((((x+,y+)<1>,z)<2>,B)<3>,A)<4>,C)<5>,(x*,y*)<6>)<7>,D)<8>
2 x D 8949 ((((((x+,y)<1>,z)<2>,B)<3>,A)<4>,C)<5>,(D,x*)<6>)<7>
1 x C 9109 ((((((x+,y)<1>,z)<2>,B)<3>,A)<4>,(C,x*)<5>)<6>,D)<7>
8 <1> <6> 9202 (((((((x+,y+)<1>,z)<2>,B)<3>,A)<4>,C)<5>,D)<6>,(x*,y*)<7>)<8>
3 x <5> 9259 (((((((x+,y)<1>,z)<2>,B)<3>,A)<4>,C)<5>,x*)<6>,D)<7>
4 x <6> 9304 (((((((x+,y)<1>,z)<2>,B)<3>,A)<4>,C)<5>,D)<6>,x*)<7>
GRAMPA tells us MUL-tree 9 is the lowest scoring tree:
((((((x+,y+)<1>,z+)<2>,B)<3>,A)<4>,(C,((x*,y*)<5>,z*)<6>)<7>)<8>,D)<9>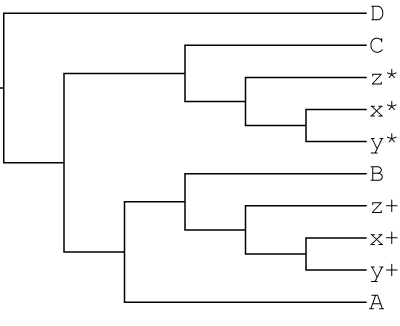
In other words, GRAMPA has identified the x,y,z clade as the polyploid clade and has identified the C lineage as the second parental lineage!
